Run Cellpose with appose#
Now let’s do something more exciting - run Cellpose segmentation using Appose! This shows how to send real data (an image) to another environment and get meaningful results back.
Important: Set your notebook kernel to ‘Python (appose_napari_ai)’ - this environment doesn’t have Cellpose installed, which perfectly demonstrates calling Cellpose from a different environment!
Load Libraries and Check Local Environment#
First, let’s import what we need. Notice we check if cellpose is available locally - it probably isn’t, and that’s okay! We’ll run it in another environment.
from skimage.io import imread
from matplotlib import pyplot as plt
import appose
try:
import cellpose
except ImportError:
print("Cellpose is not installed.")
Cellpose is not installed.
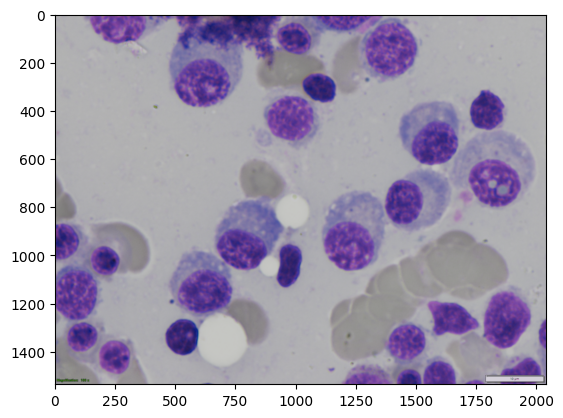
Load Our Test Image#
Let’s load the same cell image we’ve been using. This will be sent to the Cellpose environment for segmentation.
image_path = r"..\..\data\SOTA_segmentation\cell_00068.bmp"
img = imread(image_path)
plt.imshow(img)
<matplotlib.image.AxesImage at 0x1d06f8a6810>
Write the Cellpose Code#
We write Cellpose code as a string that will run in the other environment. Note how we get the result back through task.outputs.
# Create the execution string
execution_string = f'''
import numpy as np
from cellpose import models
import appose
model = models.CellposeModel(gpu=True)
masks, flows, styles = model.eval(
image.ndarray(),
#diameter=30,
flow_threshold=0.5,
niter=200
)
# Convert result to appose format for output
ndarr_mask = appose.NDArray(dtype=str(masks.dtype), shape=masks.shape)
ndarr_mask.ndarray()[:] = masks
task.outputs["mask"] = ndarr_mask
'''
Prepare the Image for Sending#
We need to convert our image into Appose’s format so it can be sent to the other environment.
cellpose_env_path = r"..\..\pixi\microsam_cellposesam\.pixi\envs\default"
env = appose.base(cellpose_env_path).build()
ndarr_img = appose.NDArray(dtype=str(img.dtype), shape=img.shape)
ndarr_img.ndarray()[:] = img
# Always include image, optionally add additional inputs
inputs = {"image": ndarr_img}
Run Cellpose in the Other Environment#
Now we send our image and code to the Cellpose environment, wait for it to finish, and get our segmentation mask back.
with env.python() as python:
task = python.task(execution_string, inputs=inputs, queue="main")
task.wait_for()
if task.error:
print(f"⚠️ Task error: {task.error}")
print(task.outputs.keys())
result = task.outputs.get("mask", None)
⚠️ Task error: Traceback (most recent call last):
File "c:\Users\bnort\work\ImageJ2022\tnia\notebooks-and-napari-widgets-for-dl\pixi\microsam_cellposesam\.pixi\envs\default\Lib\site-packages\appose\python_worker.py", line 145, in _run
exec(compile(block, "<string>", mode="exec"), _globals, binding)
File "<string>", line 8, in <module>
File "c:\Users\bnort\work\ImageJ2022\tnia\notebooks-and-napari-widgets-for-dl\pixi\microsam_cellposesam\.pixi\envs\default\Lib\site-packages\cellpose\models.py", line 338, in eval
masks = self._compute_masks(x.shape, dP, cellprob, flow_threshold=flow_threshold,
^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^
File "c:\Users\bnort\work\ImageJ2022\tnia\notebooks-and-napari-widgets-for-dl\pixi\microsam_cellposesam\.pixi\envs\default\Lib\site-packages\cellpose\models.py", line 524, in _compute_masks
outputs = dynamics.resize_and_compute_masks(
^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^
File "c:\Users\bnort\work\ImageJ2022\tnia\notebooks-and-napari-widgets-for-dl\pixi\microsam_cellposesam\.pixi\envs\default\Lib\site-packages\cellpose\dynamics.py", line 610, in resize_and_compute_masks
mask = compute_masks(dP, cellprob, niter=niter,
^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^
File "c:\Users\bnort\work\ImageJ2022\tnia\notebooks-and-napari-widgets-for-dl\pixi\microsam_cellposesam\.pixi\envs\default\Lib\site-packages\cellpose\dynamics.py", line 672, in compute_masks
mask = remove_bad_flow_masks(mask, dP, threshold=flow_threshold,
^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^
File "c:\Users\bnort\work\ImageJ2022\tnia\notebooks-and-napari-widgets-for-dl\pixi\microsam_cellposesam\.pixi\envs\default\Lib\site-packages\cellpose\dynamics.py", line 443, in remove_bad_flow_masks
merrors, _ = flow_error(masks, flows, device0)
^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^
File "c:\Users\bnort\work\ImageJ2022\tnia\notebooks-and-napari-widgets-for-dl\pixi\microsam_cellposesam\.pixi\envs\default\Lib\site-packages\cellpose\dynamics.py", line 300, in flow_error
dP_masks = masks_to_flows_gpu(maski, device=device)
^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^
File "c:\Users\bnort\work\ImageJ2022\tnia\notebooks-and-napari-widgets-for-dl\pixi\microsam_cellposesam\.pixi\envs\default\Lib\site-packages\cellpose\dynamics.py", line 139, in masks_to_flows_gpu
mu = _extend_centers_gpu(neighbors, meds_p, isneighbor, shape, n_iter=n_iter,
^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^
File "c:\Users\bnort\work\ImageJ2022\tnia\notebooks-and-napari-widgets-for-dl\pixi\microsam_cellposesam\.pixi\envs\default\Lib\site-packages\cellpose\dynamics.py", line 45, in _extend_centers_gpu
Tneigh *= isneighbor
RuntimeError: CUDA error: an illegal memory access was encountered
CUDA kernel errors might be asynchronously reported at some other API call, so the stacktrace below might be incorrect.
For debugging consider passing CUDA_LAUNCH_BLOCKING=1
Compile with `TORCH_USE_CUDA_DSA` to enable device-side assertions.
dict_keys([])
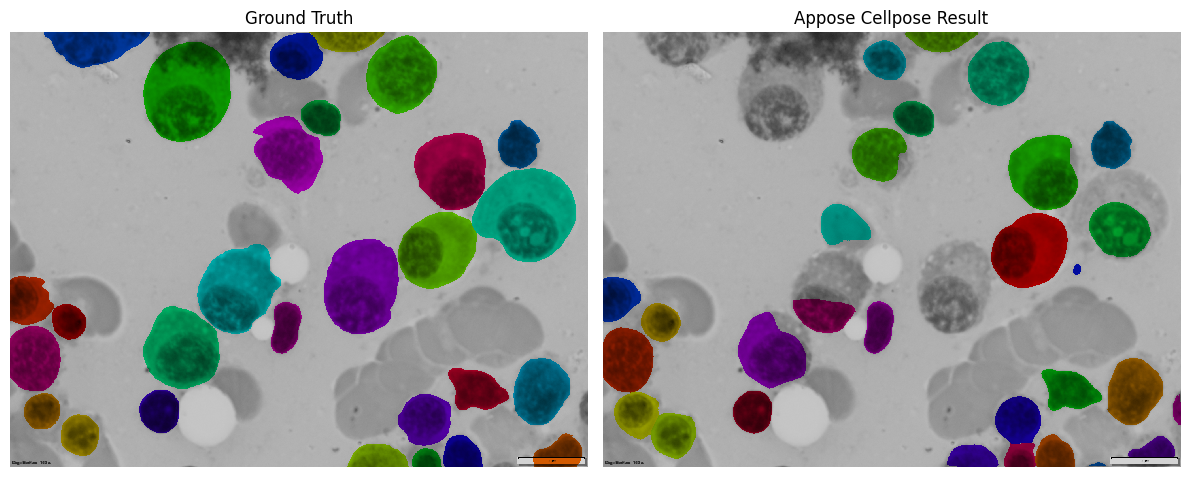
Compare Results#
Let’s see how well our Appose-powered Cellpose did! We’ll compare it side-by-side with the ground truth.
# Load ground truth for comparison
label_path = r"..\..\data\SOTA_segmentation\cell_00068_label.tiff"
ground_truth = imread(label_path)
# Import visualization helper
from tnia.plotting.plt_helper import mask_overlay
# Create overlays
ground_truth_overlay = mask_overlay(img, ground_truth)
result_overlay = mask_overlay(img, result.ndarray())
# Create comparison plot
fig, axes = plt.subplots(1, 2, figsize=(12, 6))
axes[0].imshow(ground_truth_overlay)
axes[0].set_title('Ground Truth')
axes[0].axis('off')
axes[1].imshow(result_overlay)
axes[1].set_title('Appose Cellpose Result')
axes[1].axis('off')
plt.tight_layout()
plt.show()