Easy-Augment-Batch-DL with Cell Data#
This notebook demonstrates how to use the napari-easy-augment-batch-dl plugin to train deep learning models on cell segmentation data.
Background#
The easy-augment-batch-dl plugin provides a streamlined workflow for training deep learning models with minimal data. It has panes that allow you to
Draw labels
Augment labels
Train/predict with different architectures
Semantic vs Instance Segmentation#
Semantic Segmentation: Every pixel gets classified into a category (background, vessel, cell, etc.) Instance Segmentation: Individual objects are separated (cell 1, cell 2, cell 3, etc.)
For this cell example, we’ll use instance segmentation to identify individual cells.
Labeling Strategy#
Sparse Labeling: Not every pixel has to be labeled. However some background needs to be labeled to differentiate between background pixels and unlabeled pixels. For example if there were 2 foreground classes use label 1 for background, 2 for “class 1”, and 3 for “class 2”. Unlabeled pixels (0) will be ignored.
Dense Labeling: Every foreground pixel needs to be labeled however background does not need to be labeled. If there were 2 foreground classes, use 1 for “class 1”, 2 for “class 2” and then the remaining pixels (0) will be treated as background.
In this example we use dense labeling, as we label every object in the ROI.
Augmentation#
The plugin includes a panel with various augmentation options. Simply check the desired augmentation types (Horizontal Flip, Random Resize, Random Adjust Color, Elastic Deformation, etc.) and click “Augment All Images” or “Augment Single” to automatically generate additional training data from your labeled images.
Train and Predict#
After generating augmented patches, you can train a model. Use the dropdown menu to select your desired architecture (in the screenshot, Micro-sam Instance Framework is selected), then click “Train Network”. Once training is complete, you can use your trained model to predict on new images. You also have the option to load a previously trained model if available.
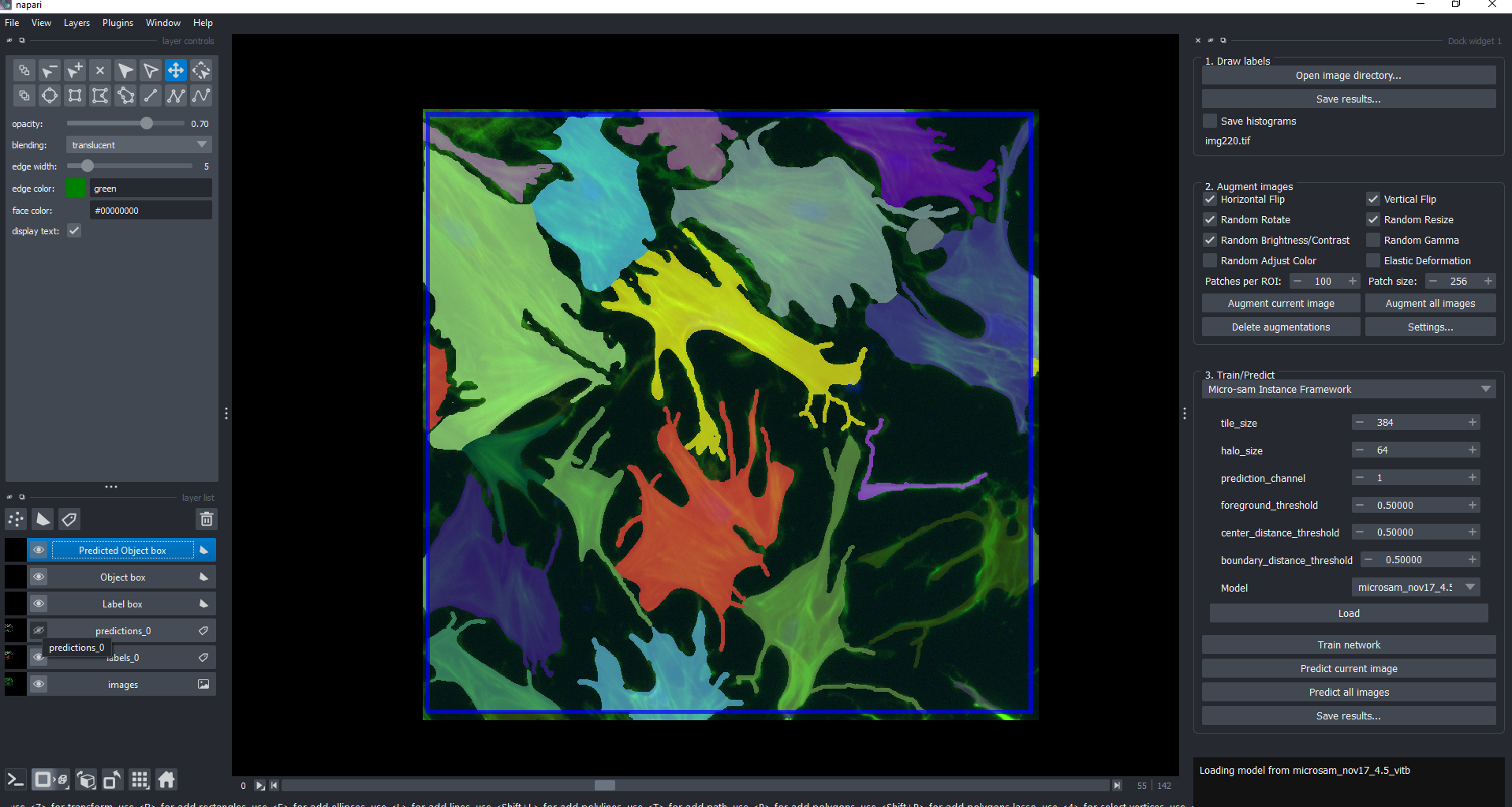
The below screenshot shows Napari-Easy-Augment-Batch open with Instance labels for individual cells drawn.
Explore Data#
This notebook uses the napari-easy-augment-batch-dl widget to explore and label the data. If we have a model trained we can also predict using that model.
Note: napari-easy-augment-batch-dl is a useful tool, especially for labelling, but is currently under construction for other uses. Right now it may be best to use it for labelling and inspecting predictions and do other steps of the deep learning workflow (making patches, training) in notebooks. (of course you are welcome to try the GUI for other steps and report and hiccups (or disasters) that occur)
from cellpose import models, io
import os
import numpy as np
import napari
from napari_easy_augment_batch_dl import easy_augment_batch_dl
from napari_easy_augment_batch_dl.frameworks.micro_sam_instance_framework import MicroSamInstanceFramework
raster_geometry not imported. This is only needed for the ellipsoid rendering in apply_stardist
c:\Users\bnort\work\ImageJ2022\tnia\notebooks-and-napari-widgets-for-dl\pixi\microsam_cellposesam\.pixi\envs\default\Lib\site-packages\tqdm\auto.py:21: TqdmWarning: IProgress not found. Please update jupyter and ipywidgets. See https://ipywidgets.readthedocs.io/en/stable/user_install.html
from .autonotebook import tqdm as notebook_tqdm
No module named 'segment_everything'
viewer = napari.Viewer()
batch_dl = easy_augment_batch_dl.NapariEasyAugmentBatchDL(viewer, label_only = False)
viewer.window.add_dock_widget(
batch_dl
)
parent_path = r'..\..\data\cells_with_protrusions'
model_path = os.path.join(parent_path, 'models', 'checkpoints')
framework_type = "Micro-sam Instance Framework"
batch_dl.load_image_directory(parent_path)
model_name = None #"microsam_nov2025_3.5_vitb"
# optionally set a pretrained model and settings so we can do prediction
batch_dl.network_architecture_drop_down.setCurrentText(framework_type)
model = batch_dl.deep_learning_project.frameworks[framework_type]
if model_name is not None:
model.model_name = model_name
widget = batch_dl.deep_learning_widgets[framework_type]
widget.sync_with_framework()
C:\Users\bnort\AppData\Local\Temp\ipykernel_40528\3280710511.py:2: DeprecationWarning: The 'label_only' parameter is deprecated. Please use the 'mode' parameter instead.
batch_dl = easy_augment_batch_dl.NapariEasyAugmentBatchDL(viewer, label_only = False)
No module named 'segment_everything'
Found framework MicroSamInstanceFramework
Found framework CellPoseInstanceFramework
creating new log file
2025-11-18 19:59:58,551 [INFO] WRITING LOG OUTPUT TO C:\Users\bnort\.cellpose\run.log
2025-11-18 19:59:58,552 [INFO]
cellpose version: 4.0.7
platform: win32
python version: 3.11.14
torch version: 2.6.0
2025-11-18 19:59:58,553 [WARNING] model_type argument is not used in v4.0.1+. Ignoring this argument...
2025-11-18 19:59:58,735 [INFO] ** TORCH CUDA version installed and working. **
2025-11-18 19:59:58,737 [INFO] >>>> using GPU (CUDA)
2025-11-18 20:00:00,673 [INFO] >>>> loading model C:\Users\bnort\.cellpose\models\cpsam
Found framework RandomForestFramework
Found framework VesselsSemanticFramework
Error creating ml labels and features: No Zarr data type found that matches {'name': '|O', 'object_codec_id': 'json2'}
Error creating ml_labels: 'DeepLearningProject' object has no attribute 'ml_labels'
Random Forest ML may not work properly
Adding object boxes layer
Adding predicted object boxes layer
Adding label boxes
Data changed
Data changed
Adding object boxes
Adding predicted object boxes
Setting object box classes
Setting predicted object box classes
