Finetuning Segment Anything with µsam#
This notebook shows how to use Segment Anything for Microscopy to fine-tune a Segment Anything Model (SAM) on the images of cells with long protrusions from this question.
(this is a derivative of the micro-sam fine tuning example)
Running this notebook with Pixi generated environment#
If you have an environment with µsam on your computer you can run this notebook in there. You can follow the installation instructions to install it on your computer.
You can also run this notebook in the cloud on Kaggle Notebooks. This service offers free usage of a GPU to speed up running the code. The next cells will take care of the installation for you if you are using it.
import torch
import os
print(torch.cuda.is_available()) # Returns True if a GPU is available
print(torch.cuda.current_device()) # Returns the current GPU device index
print(torch.cuda.device_count()) # Number of available GPUs
print(torch.cuda.get_device_name(0)) # Name of the first GPU (if available)
True
0
1
NVIDIA GeForce RTX 3090
Importing the libraries#
import warnings
import os
from tnia.plotting.plt_helper import random_label_cmap
warnings.filterwarnings("ignore")
import micro_sam
print(f"Using micro_sam version {micro_sam.__version__}")
from glob import glob
from IPython.display import FileLink
import numpy as np
import imageio.v3 as imageio
from matplotlib import pyplot as plt
from skimage.measure import label as connected_components
import torch
from torch_em.util.debug import check_loader
from torch_em.data import MinInstanceSampler
from torch_em.util.util import get_random_colors
import micro_sam.training as sam_training
from micro_sam.sample_data import fetch_tracking_example_data, fetch_tracking_segmentation_data
from micro_sam.automatic_segmentation import get_predictor_and_segmenter, automatic_instance_segmentation
Using micro_sam version 1.6.2
Set paths#
parent_path = r'..\..\data\cells_with_protrusions'
train_path = os.path.join(parent_path, 'patches')
model_path = os.path.join(parent_path, 'models')
segmentation_dir = train_path + '/ground truth0'
image_dir = train_path + '/input0'
convert to 8 bit#
For micro-sam inputs must be in 8 bit.
image_dir_255 = train_path + '/input0_255'
# if no 8-bit images yet make them
if not os.path.exists(image_dir_255):
os.makedirs(image_dir_255)
image_paths = sorted(glob(os.path.join(image_dir, "*")))
for image_path in image_paths:
image = imageio.imread(image_path)
image = (image * 255).astype('uint8')
imageio.imwrite(image_path.replace(image_dir, image_dir_255), image)
Let’s create the dataloaders#
Our task is to segment HeLa cells on a flat glass in DIC microscopic images. The dataset comes from https://celltrackingchallenge.net/2d-datasets/, and the dataloader has been implemented in torch-em.
First, let’s visualize how our samples look.#
image_dir = image_dir_255
image_paths = sorted(glob(os.path.join(image_dir, "*")))
print('num images is', len(image_paths))
segmentation_paths = sorted(glob(os.path.join(segmentation_dir, "*")))
print('num segmentations is', len(segmentation_paths))
for image_path, segmentation_path in zip(image_paths, segmentation_paths):
image = imageio.imread(image_path)
print('image size is', image.shape)
segmentation = imageio.imread(segmentation_path)
fig, ax = plt.subplots(1, 2, figsize=(10, 10))
ax[0].imshow(image, cmap="gray")
ax[0].set_title("Input Image")
ax[0].axis("off")
segmentation = connected_components(segmentation)
ax[1].imshow(segmentation, cmap=random_label_cmap(), interpolation="nearest")
ax[1].set_title("Ground Truth Instances")
ax[1].axis("off")
plt.show()
plt.close()
break # comment this out in case you want to visualize all the images
image_paths_to_remove = []
segmentation_paths_to_remove = []
for image_path, segmentation_path in zip(image_paths, segmentation_paths):
segmentation = imageio.imread(segmentation_path)
if segmentation.max() == 0:
print(f"Warning: segmentation {segmentation_path} is empty.")
image_paths_to_remove.append(image_path)
segmentation_paths_to_remove.append(segmentation_path)
print()
print(f"Removing {len(image_paths_to_remove)} images with empty segmentations.")
for image_path, segmentation_path in zip(image_paths_to_remove, segmentation_paths_to_remove):
# remove empty file (because microsam throws an error if empty segmentation is present)
print(f"Removing {image_path} and {segmentation_path}")
os.remove(image_path)
os.remove(segmentation_path)
num images is 360
num segmentations is 360
image size is (512, 512, 3)
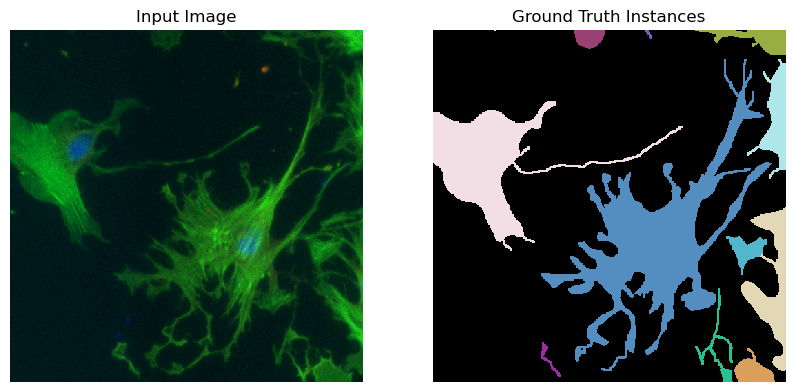
Removing 0 images with empty segmentations.
# 'micro_sam.training.default_sam_loader' is a convenience function to build a pytorch dataloader from image data and labels for training segmentation models.
# This is wrapped around the 'torch_em.default_segmentation_loader'.
# It supports image data in various formats.
# Here, we load image data and labels from the two folders with tif images that were downloaded by the example data functionality,
# by specifying `raw_key` and `label_key` as `*.tif`.
# This means all images in the respective folders that end with .tif will be loaded.
# The function supports many other file formats. For example, if you have tif stacks with multiple slices instead of multiple tif images in a folder,
# then you can pass 'raw_key=label_key=None'.
# For more information, here is the documentation: https://github.com/constantinpape/torch-em/blob/main/torch_em/data/datasets/README.md
# And here is a tutorial on creating dataloaders using 'torch-em': https://github.com/constantinpape/torch-em/blob/main/notebooks/tutorial_create_dataloaders.ipynb
# Load images from multiple files in folder via pattern (here: all tif files)
raw_key, label_key = "*.tif", "*.tif"
# Alternative: if you have tif stacks you can just set 'raw_key' and 'label_key' to None
# raw_key, label_key= None, None
# The 'roi' argument can be used to subselect parts of the data.
# Here, we use it to select the first 70 images (frames) for the train split and the other frames for the val split.
train_roi = np.s_[:1180, :, :]
val_roi = np.s_[1180:1200, :, :]
# The script below returns the train or val data loader for finetuning Segment Anything Model (SAM).
# The data loader must be a torch data loader that returns `x, y` tensors, where `x` is the image data and `y` are the labels.
# The labels have to be in a label mask instance segmentation format.
# i.e. a tensor of the same spatial shape as `x`, with each object mask having its own ID.
# Important: the ID 0 is reseved for background, and the IDs must be consecutive
# Here, we use `micro_sam.training.default_sam_loader` for creating a suitable data loader from
# the example hela data. You can either adapt this for your own data or write a suitable torch dataloader yourself.
# Here's a quickstart notebook to create your own dataloaders: https://github.com/constantinpape/torch-em/blob/main/notebooks/tutorial_create_dataloaders.ipynb
batch_size = 1 # the training batch size
patch_shape = (1, 512, 512) # the size of patches for training
# Train an additional convolutional decoder for end-to-end automatic instance segmentation
# NOTE 1: It's important to have densely annotated-labels while training the additional convolutional decoder.
# NOTE 2: In case you do not have labeled images, we recommend using `micro-sam` annotator tools to annotate as many objects as possible per image for best performance.
train_instance_segmentation = True
# NOTE: The dataloader internally takes care of adding label transforms: i.e. used to convert the ground-truth
# labels to the desired instances for finetuning Segment Anythhing, or, to learn the foreground and distances
# to the object centers and object boundaries for automatic segmentation.
# There are cases where our inputs are large and the labeled objects are not evenly distributed across the image.
# For this we use samplers, which ensure that valid inputs are chosen subjected to the paired labels.
# The sampler chosen below makes sure that the chosen inputs have atleast one foreground instance, and filters out small objects.
sampler = MinInstanceSampler(min_size=1) # NOTE: The choice of 'min_size' value is paired with the same value in 'min_size' filter in 'label_transform'.
train_loader = sam_training.default_sam_loader(
raw_paths=image_dir,
raw_key=raw_key,
label_paths=segmentation_dir,
label_key=label_key,
with_segmentation_decoder=train_instance_segmentation,
patch_shape=patch_shape,
batch_size=batch_size,
is_seg_dataset=True,
rois=None,#train_roi,
shuffle=True,
raw_transform=sam_training.identity,
sampler=sampler,
#with_channels = True,
)
val_loader = sam_training.default_sam_loader(
raw_paths=image_dir,
raw_key=raw_key,
label_paths=segmentation_dir,
label_key=label_key,
with_segmentation_decoder=train_instance_segmentation,
patch_shape=patch_shape,
batch_size=batch_size,
is_seg_dataset=True,
rois=None, #val_roi,
shuffle=True,
raw_transform=sam_training.identity,
sampler=sampler,
#with_channels = True,
)
..\..\data\cells_with_protrusions\patches/input0_255 ..\..\data\cells_with_protrusions\patches/ground truth0 None
..\..\data\cells_with_protrusions\patches/input0_255 ..\..\data\cells_with_protrusions\patches/ground truth0 None
..\..\data\cells_with_protrusions\patches/input0_255 ..\..\data\cells_with_protrusions\patches/ground truth0 None
..\..\data\cells_with_protrusions\patches/input0_255 ..\..\data\cells_with_protrusions\patches/ground truth0 None
test = train_loader.dataset[0]
test[0].shape, test[1].shape
test[0].min(), test[0].max(), test[1].min(), test[1].max()
(tensor(0.), tensor(255.), tensor(0.), tensor(22.))
# Let's check how our samples look from the dataloader
check_loader(train_loader, 5, plt=True)
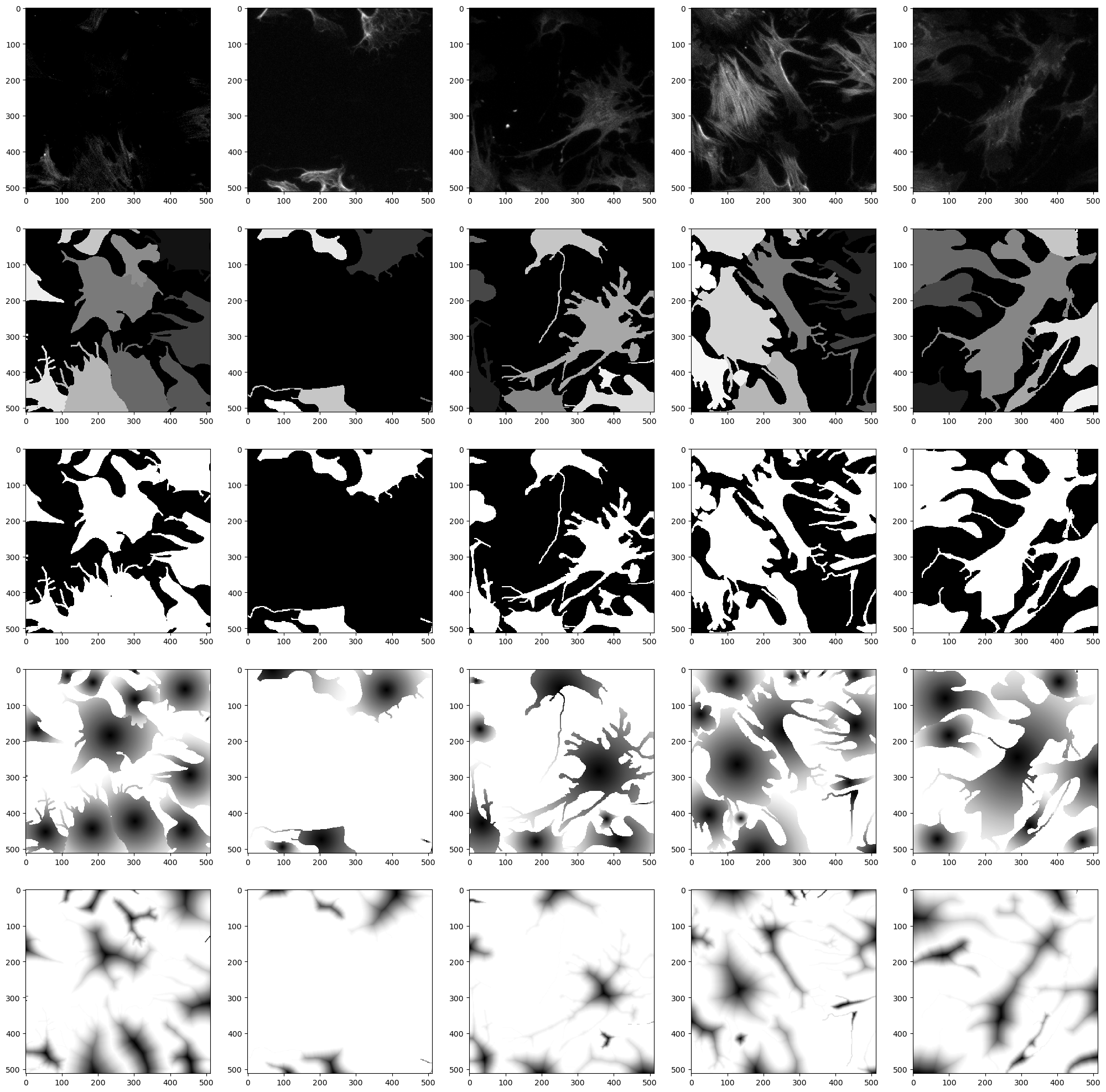
Run the actual model finetuning#
# All hyperparameters for training.
n_objects_per_batch = 2 # the number of objects per batch that will be sampled
device = "cuda" if torch.cuda.is_available() else "cpu" # the device/GPU used for training
n_epochs = 5 # how long we train (in epochs)
# The model_type determines which base model is used to initialize the weights that are finetuned.
# We use vit_b here because it can be trained faster. Note that vit_h usually yields higher quality results.
model_type = "vit_b"
# The name of the checkpoint. The checkpoints will be stored in './checkpoints/<checkpoint_name>'
checkpoint_name = "microsam_nov17_4.5_vitb"
NOTE: The user needs to decide whether to finetune the Segment Anything model, or the µsam’s “finetuned microscopy models” for their dataset. Here, we finetune on the Segment Anything model for simplicity. For example, if you choose to finetune the model from the light microscopy generalist models, you need to update the model_type to vit_b_lm and it takes care of initializing the model with the desired weights)
help(sam_training.train_sam)
Help on function train_sam in module micro_sam.training.training:
train_sam(name: str, model_type: str, train_loader: torch.utils.data.dataloader.DataLoader, val_loader: torch.utils.data.dataloader.DataLoader, n_epochs: int = 100, early_stopping: Optional[int] = 10, n_objects_per_batch: Optional[int] = 25, checkpoint_path: Union[str, os.PathLike, NoneType] = None, with_segmentation_decoder: bool = True, freeze: Optional[List[str]] = None, device: Union[str, torch.device, NoneType] = None, lr: float = 1e-05, n_sub_iteration: int = 8, save_root: Union[str, os.PathLike, NoneType] = None, mask_prob: float = 0.5, n_iterations: Optional[int] = None, scheduler_class: Optional[torch.optim.lr_scheduler._LRScheduler] = <class 'torch.optim.lr_scheduler.ReduceLROnPlateau'>, scheduler_kwargs: Optional[Dict[str, Any]] = None, save_every_kth_epoch: Optional[int] = None, pbar_signals: Optional[PyQt5.QtCore.QObject] = None, optimizer_class: Optional[torch.optim.optimizer.Optimizer] = <class 'torch.optim.adamw.AdamW'>, peft_kwargs: Optional[Dict] = None, ignore_warnings: bool = True, verify_n_labels_in_loader: Optional[int] = 50, box_distortion_factor: Optional[float] = 0.025, overwrite_training: bool = True, **model_kwargs) -> None
Run training for a SAM model.
Args:
name: The name of the model to be trained. The checkpoint and logs will have this name.
model_type: The type of the SAM model.
train_loader: The dataloader for training.
val_loader: The dataloader for validation.
n_epochs: The number of epochs to train for.
early_stopping: Enable early stopping after this number of epochs without improvement.
By default, the value is set to '10' epochs.
n_objects_per_batch: The number of objects per batch used to compute
the loss for interative segmentation. If None all objects will be used,
if given objects will be randomly sub-sampled. By default, the number of objects per batch are '25'.
checkpoint_path: Path to checkpoint for initializing the SAM model.
with_segmentation_decoder: Whether to train additional UNETR decoder for automatic instance segmentation.
By default, trains with the additional instance segmentation decoder.
freeze: Specify parts of the model that should be frozen, namely: image_encoder, prompt_encoder and mask_decoder
By default nothing is frozen and the full model is updated.
device: The device to use for training. By default, automatically chooses the best available device to train.
lr: The learning rate. By default, set to '1e-5'.
n_sub_iteration: The number of iterative prompts per training iteration.
By default, the number of iterations is set to '8'.
save_root: Optional root directory for saving the checkpoints and logs.
If not given the current working directory is used.
mask_prob: The probability for using a mask as input in a given training sub-iteration.
By default, set to '0.5'.
n_iterations: The number of iterations to use for training. This will over-ride `n_epochs` if given.
scheduler_class: The learning rate scheduler to update the learning rate.
By default, `torch.optim.lr_scheduler.ReduceLROnPlateau` is used.
scheduler_kwargs: The learning rate scheduler parameters.
If passed 'None', the chosen default parameters are used in `ReduceLROnPlateau`.
save_every_kth_epoch: Save checkpoints after every kth epoch separately.
pbar_signals: Controls for napari progress bar.
optimizer_class: The optimizer class. By default, `torch.optim.AdamW` is used.
peft_kwargs: Keyword arguments for the PEFT wrapper class.
ignore_warnings: Whether to ignore raised warnings. By default, set to 'True'.
verify_n_labels_in_loader: The number of labels to verify out of the train and validation dataloaders.
By default, 50 batches of labels are verified from the dataloaders.
box_distortion_factor: The factor for distorting the box annotations derived from the ground-truth masks.
By default, the distortion factor is set to '0.025'.
overwrite_training: Whether to overwrite the trained model stored at the same location.
By default, overwrites the trained model at each run.
If set to 'False', it will avoid retraining the model if the previous run was completed.
model_kwargs: Additional keyword arguments for the `micro_sam.util.get_sam_model`.
# Run training (best metric 0.027211)
sam_training.train_sam(
name=checkpoint_name,
save_root=model_path, #os.path.join(root_dir, "models"),
model_type=model_type,
train_loader=train_loader,
val_loader=val_loader,
n_epochs=n_epochs,
n_objects_per_batch=n_objects_per_batch,
with_segmentation_decoder=train_instance_segmentation,
device=device,
)
Verifying labels in 'train' dataloader: 100%|██████████| 50/50 [00:05<00:00, 8.90it/s]
Verifying labels in 'val' dataloader: 100%|██████████| 50/50 [00:05<00:00, 9.14it/s]
Start fitting for 1800 iterations / 5 epochs
with 360 iterations per epoch
Training with mixed precision
Epoch 5: average [s/it]: 0.928116, current metric: 0.176352, best metric: 0.176352: 100%|█████████▉| 1799/1800 [38:50<00:01, 1.30s/it]
Finished training after 5 epochs / 1800 iterations.
The best epoch is number 4.
Training took 2343.2302627563477 seconds (= 00:39:03 hours)
The Kernel crashed while executing code in the current cell or a previous cell.
Please review the code in the cell(s) to identify a possible cause of the failure.
Click <a href='https://aka.ms/vscodeJupyterKernelCrash'>here</a> for more info.
View Jupyter <a href='command:jupyter.viewOutput'>log</a> for further details.
# Let's spot our best checkpoint and download it to get started with the annotation tool
best_checkpoint = os.path.join(model_path, "checkpoints", checkpoint_name, "best.pt")
# Download link is automatically generated for the best model.
print("Click here \u2193")
FileLink(best_checkpoint)
Let’s run the automatic instance segmentation (AIS)#
def run_automatic_instance_segmentation(image, checkpoint_path, model_type="vit_b_lm", device=None, tile_shape = None, halo = None):
"""Automatic Instance Segmentation (AIS) by training an additional instance decoder in SAM.
NOTE: AIS is supported only for `µsam` models.
Args:
image: The input image.
checkpoint_path: The path to stored checkpoints.
model_type: The choice of the `µsam` model.
device: The device to run the model inference.
Returns:
The instance segmentation.
"""
# Step 1: Get the 'predictor' and 'segmenter' to perform automatic instance segmentation.
predictor, segmenter = get_predictor_and_segmenter(
model_type=model_type, # choice of the Segment Anything model
checkpoint=checkpoint_path, # overwrite to pass your own finetuned model.
device=device, # the device to run the model inference.
is_tiled = (tile_shape is not None), # whether the model is tiled or not.
)
# Step 2: Get the instance segmentation for the given image.
prediction = automatic_instance_segmentation(
predictor=predictor, # the predictor for the Segment Anything model.
segmenter=segmenter, # the segmenter class responsible for generating predictions.
input_path=image,
ndim=2,
tile_shape=tile_shape,
halo=halo,
)
return prediction
assert os.path.exists(best_checkpoint), "Please train the model first to run inference on the finetuned model."
assert train_instance_segmentation is True, "Oops. You didn't opt for finetuning using the decoder-based automatic instance segmentation."
# Let's check the first 5 images. Feel free to comment out the line below to run inference on all images.
image_paths_ = image_paths[:5]
segmentation_paths_ = segmentation_paths[:5]
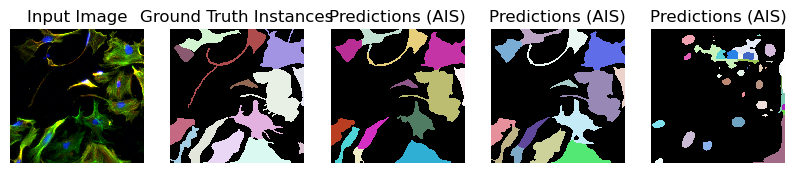
for image_path, segmentation_path in zip(image_paths_, segmentation_paths_):
image = imageio.imread(image_path)
image_ = np.transpose(image, (2, 0, 1))
segmentation = imageio.imread(segmentation_path)
# Predicted instances
prediction = run_automatic_instance_segmentation(
image=image_, checkpoint_path=best_checkpoint, model_type=model_type, device=device
)
# Visualize the predictions
fig, ax = plt.subplots(1, 5, figsize=(10, 10))
ax[0].imshow(image, cmap="gray")
ax[0].axis("off")
ax[0].set_title("Input Image")
ax[1].imshow(segmentation, cmap=random_label_cmap(), interpolation="nearest")
ax[1].axis("off")
ax[1].set_title("Ground Truth Instances")
ax[2].imshow(prediction[0], cmap=random_label_cmap(), interpolation="nearest")
ax[2].axis("off")
ax[2].set_title("Predictions (AIS)")
ax[3].imshow(prediction[1], cmap=random_label_cmap(), interpolation="nearest")
ax[3].axis("off")
ax[3].set_title("Predictions (AIS)")
ax[4].imshow(prediction[2], cmap=random_label_cmap(), interpolation="nearest")
ax[4].axis("off")
ax[4].set_title("Predictions (AIS)")
plt.show()
plt.close()
#break
Compute Image Embeddings 3D: 100%|██████████| 3/3 [00:00<00:00, 5.03it/s]
Segment slices: 100%|██████████| 3/3 [00:00<00:00, 10.00it/s]
Merge segmentation: 100%|██████████| 1/1 [00:00<00:00, 43.48it/s]
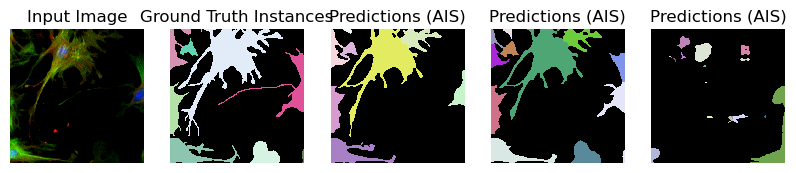
Compute Image Embeddings 3D: 100%|██████████| 3/3 [00:00<00:00, 5.32it/s]
Segment slices: 100%|██████████| 3/3 [00:00<00:00, 10.91it/s]
Merge segmentation: 100%|██████████| 1/1 [00:00<00:00, 22.22it/s]
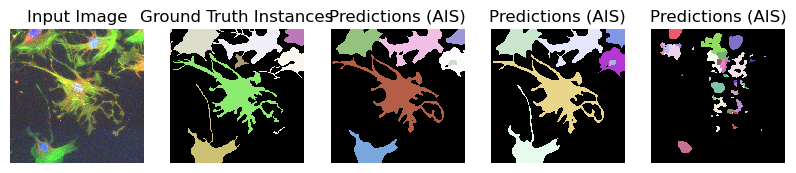
Compute Image Embeddings 3D: 100%|██████████| 3/3 [00:00<00:00, 4.67it/s]
Segment slices: 100%|██████████| 3/3 [00:00<00:00, 10.95it/s]
Merge segmentation: 100%|██████████| 1/1 [00:00<00:00, 43.49it/s]
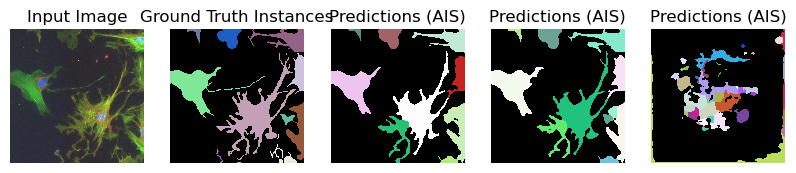
Compute Image Embeddings 3D: 100%|██████████| 3/3 [00:00<00:00, 4.75it/s]
Segment slices: 100%|██████████| 3/3 [00:00<00:00, 10.56it/s]
Merge segmentation: 100%|██████████| 1/1 [00:00<00:00, 45.45it/s]
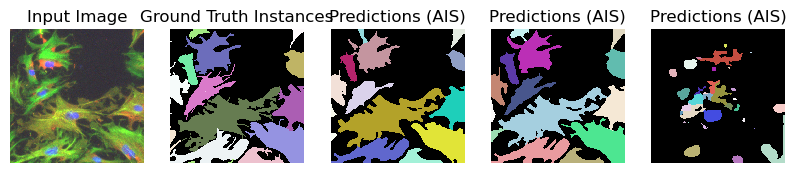
Compute Image Embeddings 3D: 100%|██████████| 3/3 [00:00<00:00, 5.19it/s]
Segment slices: 100%|██████████| 3/3 [00:00<00:00, 10.68it/s]
Merge segmentation: 100%|██████████| 1/1 [00:00<00:00, 34.48it/s]
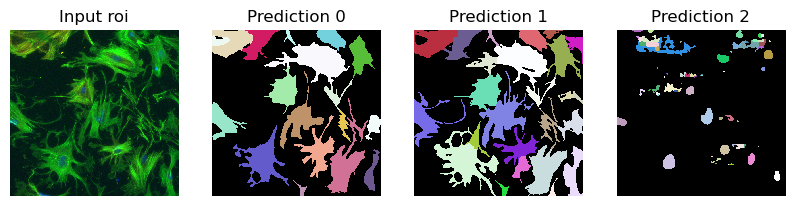
im = imageio.imread(r"D:\images\tnia-python-images\imagesc\2024_10_11_tough_cellpose_2\img52.tif")
im_ = np.transpose(im, (2, 0, 1))
prediction = run_automatic_instance_segmentation(
image=im_, checkpoint_path=best_checkpoint, model_type=model_type, device=device, tile_shape=(384, 384), halo = (64, 64)
#image=im_whole, checkpoint_path=best_checkpoint, model_type=model_type, device=device
)
# Visualize the predictions
fig, ax = plt.subplots(1, 4, figsize=(10, 10))
ax[0].imshow(im//255, cmap="gray")
ax[0].axis("off")
ax[0].set_title("Input roi")
ax[1].imshow(prediction[0], cmap=random_label_cmap(), interpolation="nearest")
ax[1].axis("off")
ax[1].set_title("Prediction 0")
ax[2].imshow(prediction[1], cmap=random_label_cmap(), interpolation="nearest")
ax[2].axis("off")
ax[2].set_title("Prediction 1")
ax[3].imshow(prediction[2], cmap=random_label_cmap(), interpolation="nearest")
ax[3].axis("off")
ax[3].set_title("Prediction 2")
plt.show()
plt.close()
Compute Image Embeddings 3D tiled: 100%|██████████| 12/12 [00:03<00:00, 3.21it/s]
Segment slices: 100%|██████████| 3/3 [00:01<00:00, 2.50it/s]
Merge segmentation: 100%|██████████| 1/1 [00:00<00:00, 20.80it/s]
WARNING:matplotlib.image:Clipping input data to the valid range for imshow with RGB data ([0..1] for floats or [0..255] for integers). Got range [0..257].
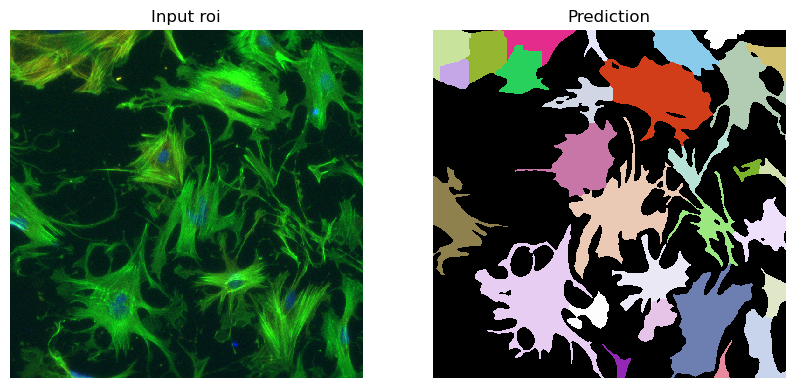
im = imageio.imread(r"D:\images\tnia-python-images\imagesc\2024_10_11_tough_cellpose_2\img52.tif")
prediction = run_automatic_instance_segmentation(
image=im, checkpoint_path=best_checkpoint, model_type=model_type, device=device, tile_shape=(384, 384), halo = (64, 64)
#image=im_whole, checkpoint_path=best_checkpoint, model_type=model_type, device=device
)
# Visualize the predictions
fig, ax = plt.subplots(1, 2, figsize=(10, 10))
ax[0].imshow(im//255, cmap="gray")
ax[0].axis("off")
ax[0].set_title("Input roi")
ax[1].imshow(prediction, cmap=random_label_cmap(), interpolation="nearest")
ax[1].axis("off")
ax[1].set_title("Prediction")
plt.show()
plt.close()
Compute Image Embeddings 2D tiled: 100%|██████████| 4/4 [00:01<00:00, 2.53it/s]
Initialize tiled instance segmentation with decoder: 100%|██████████| 4/4 [00:00<00:00, 12.12it/s]
WARNING:matplotlib.image:Clipping input data to the valid range for imshow with RGB data ([0..1] for floats or [0..255] for integers). Got range [0..257].
import napari
viewer = napari.Viewer()
viewer.add_image(im, name="Input Image", colormap="gray")
viewer.add_labels(prediction[1], name="Predictions (AIS)")
<Labels layer 'Predictions (AIS)' at 0x2906579e590>
What next?#
It’s time to get started with your custom finetuned model using the annotator tool. Here is the documentation on how to get started with µsam: Annotation Tools
Happy annotating!
This notebook was last ran on October 20, 2024
